World Placozoa Database
Table of Contents
- Introduction (The Animal)
- Biology
- Publication History
- Morphological Variation
- Phylogenetic Placement with Other Animals
- Taxonomy
- Distribution
- Bacterial Endosymbionts
- Further Milestones in Placozoan Research
- Sampling and Storage
- References
- Placozoa: A Quick Guide
- Cool (Very) Photographs and Drawings of Placozoa
I. INTRODUCTION (THE ANIMAL)
In 1883 the first species — Trichoplax adhaerens — was described and suggested to be at the base of the animal tree [1]. After a decade, Treptoplax reptans was described [2]. Unfortunately, the living T. reptans specimens from Monticelli died and have never been resampled in the field; the characteristics identified by Monticelli, however, have been doubted and this species is therefore currently unaccepted, although it could be rare or extinct. Placozoans were subsequently misclassified as adult acoel turbellarians and cnidarian larvae. Thus, decades passed without studies on Placozoa. In the 1960’s the lineage was rediscovered and soon considered distinct again. Significantly, the phylum Placozoa was formally erected [3]. More modern morphological studies have identified ultrastructural characters that differ between some placozoans, but diagnostic morphological characters remained limited [4]. This led to the view of a phylum with morphological uniformity and low taxonomic diversity. This changed dramatically in the 21st century with the use of molecular genetic techniques that led to the discrimination of different genetic lineages [5-7] and the sequencing of entire placozoan genomes [8-11]
The transition started in the early 2000s, when molecular data began to show an unexpected amount of genetic diversity in this “phylum of one” [5]. Ongoing 16S DNA sequence analyses consistently revealed new placozoan haplotypes, with genetic distances on par with higher taxa in other animal phyla. While this immense cryptic diversity became more and more obvious, the scientific community remained hesitant for over a decade to accept the formal description of animal species using only molecular markers.
In 2011 Placozoa was still considered a monotypic phylum. Here is our description of taxonomic status of the group in 2011:
“This is the smallest of all animal phyla yet it contains a single nominal species, Trichoplax adhaerens (Figure 1A). A second species that is sometimes mentioned, Treptoplax reptans, has never been found again after its first and only report and is most likely a fragmented placozoan. With only one species the phylum knows only three taxonomic ranks: Phylum (Placozoa), genus (Trichoplax), and species (T. adhaerens). This situation is soon to change radically. Genetic analyses have suggested that Placozoa harbor at least seven major species clades, which may represent different orders, each with multiple families.”
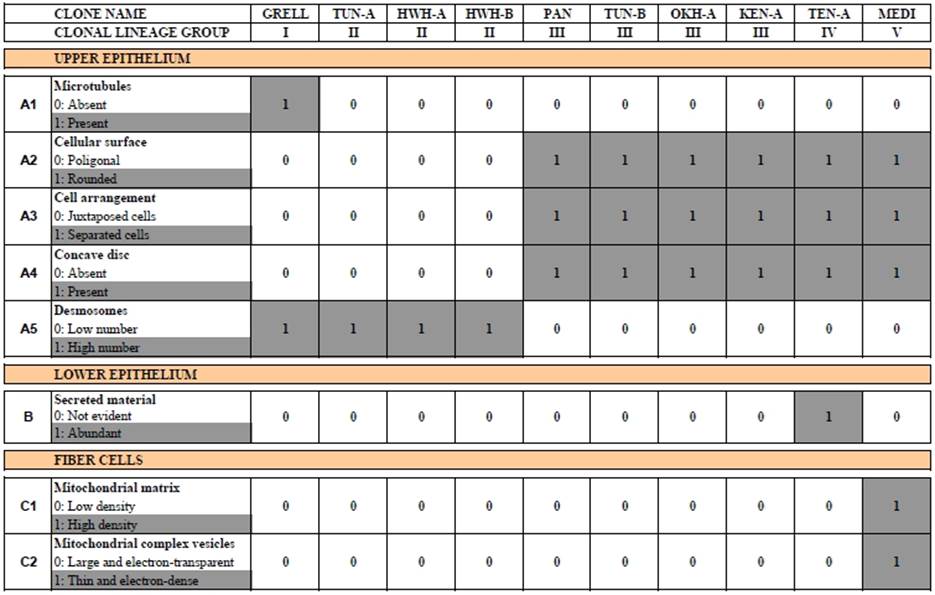
Indeed, the phylum now consists of at least two classes, four orders, several families and genera, around 30 genetically distinct lineages and four formally described species [5-7, 10, 12-15] (see current taxonomy below). A further milestone in our understanding of placozoan diversity and evolution was certainly the discovery and description of a placozoan species, Polyplacotoma mediterranea, that is morphologically different from all other placozoan species (Figure 1B) [12].
II. BIOLOGY AND HISTORY
Placozoa occur in the littoral of all warm oceans and are distributed globally in tropical and sub-tropical waters. They reproduce by (i) binary (sometimes trinary) fission, (ii) budding off small swarmers (iii) sexual reproduction.
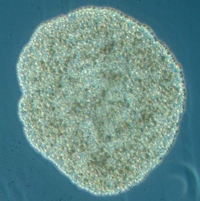
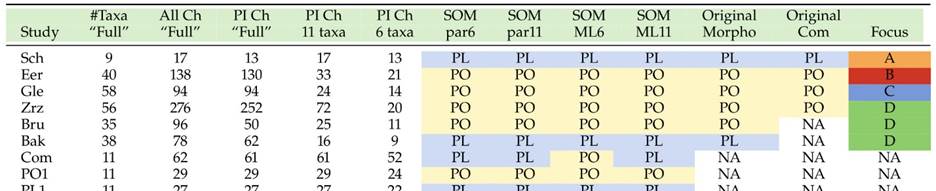
The placozoan Trichoplax adhaerens is more simply organized than any other living metazoan. This tiny marine animal, with a size of up to 2mm, looks like an irregular “hairy plate” (“tricho plax”) whose unique bauplan is based on a simple, irregular sandwich organization. An upper and a lower epithelium surround a loose network (not an epithelium) of so-called fiber cells (Fig. 2A). Traditionally only four cell types have been described in Trichoplax, upper and lower epithelial cells, gland cells within the lower, feeding epithelium, and fiber cells sandwiched between the epithelia [4, 16-19]. No organs or specialized nerve or muscular cells are present. A basal lamina and extracellular matrix are likewise lacking. All these simple bauplan characteristics make placozoans more similar to protozoans than to any other existing metazoans. Body shape is irregular and changes constantly. No symmetry of any kind is seen, and nothing like an oral-aboral or even a dorso-ventral polarity exists. All of the above justified the construction of an own phylum, Placozoa [3].
After its original description by F.E. Schulze 1883 [1], Trichoplax attracted particular attention as a potential candidate representing the basic and ancestral state of metazoan organization. The simplest of all metazoan morphologies suggested a basal position for T. adhaerens. Presently the phylogenetic position of Placozoa is subject of hot debates. For details and references on placozoan biology and the history of placozoan research see [21-23].
III. PUBLICATION HISTORY
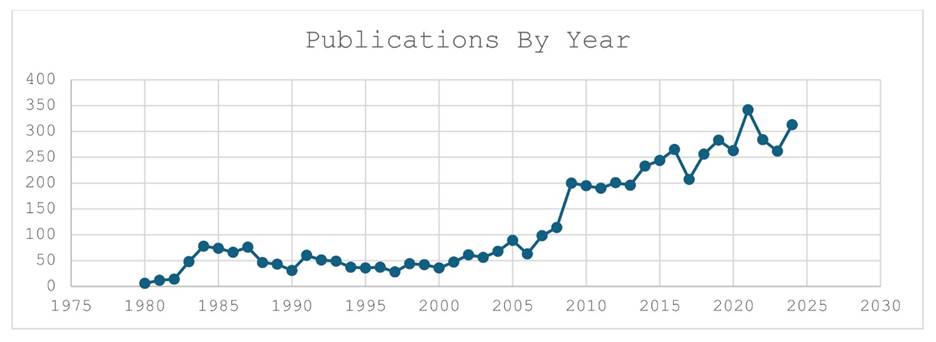
With over a century of research starting in 1883 and the publication of Schulze’s description of Trichoplax adhaerens, the phylum has been the subject of over 5560 publications. The graph shows number of publications in the Scholar database from 1980 to 2024. Between 1985 and 2005 there was a period of steady research at about 30 publications per year that we estimate came from less than ten research labs. In 2008 there was an uptick by a factor of five of papers coming from greater than fifty labs. We estimate that these papers came from over 50 different labs across the globe. The year 2008 also marked the publication of the Trichoplax adhaerens genome [8]. Since 2010 the number of papers using placozoa as a subject has stayed steady at above 200/yr with a peak in 2021 of 350 papers. This steady growth of publications indicates a healthy research community that centers on the study of this tiny animal.
IV. MORPHOLOGICAL VARIATION
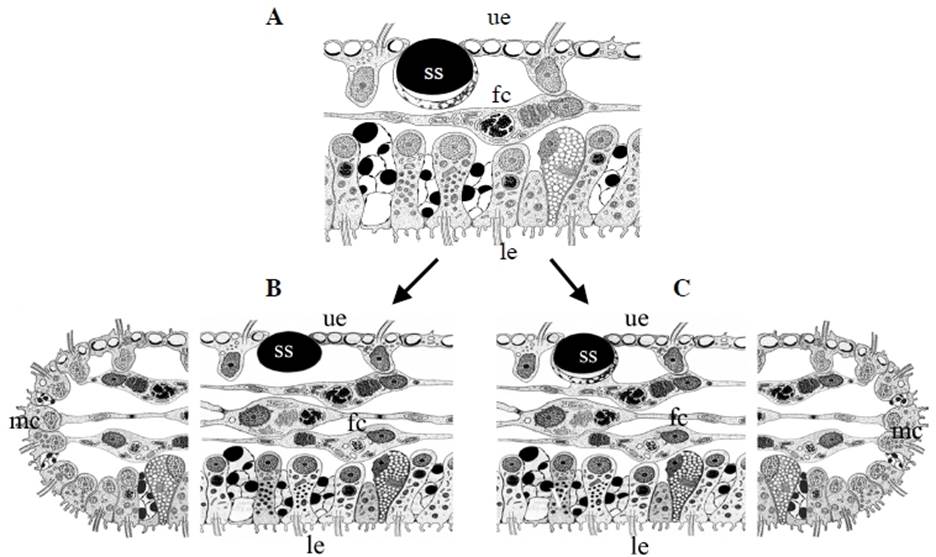
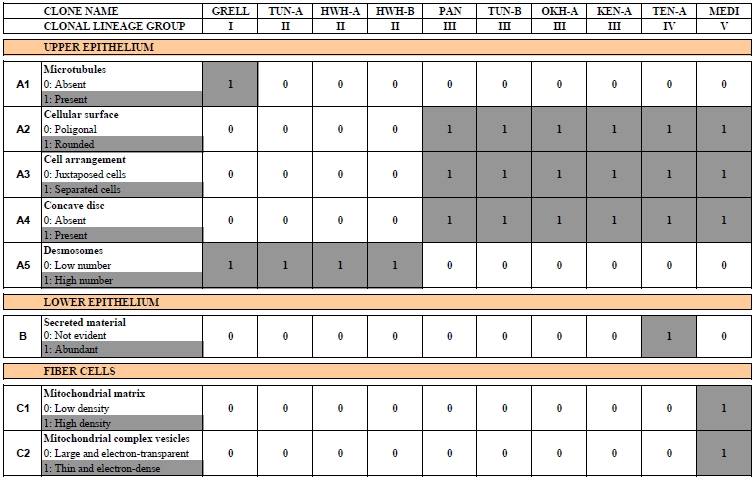
With the exception of Polyplacotoma mediterranea, placozoans are indistinguishable at the macroscopic level. However, studying genetically different placozoan lineages at the ultrastructural level revealed subtle differences (Table 1) and showed that in all lineages fiber cells are always arranged in multiple layers [20]. Trichoplax adhaerens served as a positive control in this study. Even in Trichoplax the fiber cells are, in contrast to earlier observations [16, 19], arranged in more than one layer. Fiber cells are thus organized in a 3D meshwork rather than two dimensions only (see Fig. 2B,C). Guidi et al. [20] proposed different subtypes of fiber cells. These new observations helped to better understand the nature of the placozoan bauplan.
V. PHYLOGENETIC PLACEMENT WITH OTHER ANIMALS
Taken from: Neumann, Johannes S., Rob Desalle, Apurva Narechania, Bernd Schierwater, and Michael Tessler. "Morphological characters can strongly influence early animal relationships inferred from phylogenomic data sets." Systematic biology 70, no. 2 (2021): 360-375. https://doi.org/10.1093/sysbio/syaa038 (see references therein)
We define the sister group to all other extant Metazoa as the SOM. There are only five major lineages that could be considered the SOM: Bilateria, Cnidaria, Ctenophora, Placozoa, or Porifera, allowing for 105 possible bifurcating trees. As pointed out by Schierwater et al. (2009), a large number of these topologies have appeared in publications. The hypothesis of Porifera as the SOM has prevailed since early morphological studies and into the beginning of the DNA sequence era (Field et al. 1988; Schram 1991; Backeljau 1993; Zrzavý 1998; Philippe et al. 2009). With the advent of medium-sized phylogenetic molecular data in the first decade of the 21st Century, Placozoa gained momentum as being inferred as the most likely SOM (Dellaporta et al. 2006; Signorovitch et al. 2007). As phylogenomics came into play in the early 2000s, the hypotheses switched to primarily Porifera or Ctenophora inferred as the most probable SOM (Rokas et al. 2003; Rokas and Carroll 2005; Dunn et al. 2008; Srivastava et al. 2008; Pick et al. 2010; Nesnidal et al. 2013; Ryan et al. 2013; Moroz et al. 2014; Whelan et al. 2015; Shen et al. 2017; Laumer et al. 2018).
The reasons for this disparity in hypotheses are surely varied and include sequence length differences, taxon sampling, and model application, among others. Several studies have discussed the ramifications of both Porifera and Ctenophora inferred as the SOM, and it is clear that the evolution of some of the most fundamental morphological traits in animals is at stake—neural tissue, muscle cells, and mesoderm, to name a few (see Nielsen 2019 for a recent review). For instance, if ctenophores are inferred as the SOM, then the nervous tissue and neural systems most likely evolved twice, although an equally parsimonious scenario would involve two independent losses of a nervous system, one in Porifera and one in Placozoa. If Porifera is inferred as the SOM, then the neural tissue and the nervous system almost surely evolved only once. The latter hypothesis is more intuitive and parsimonious than the former.
However, arguments as to the ease with which neural tissue and nervous systems can arise (i.e., from the neural genomic toolbox that the last common ancestor of all metazoans seems to have possessed) have also been proffered (Moroz et al. 2006; Moroz 2009, 2015). While these scenarios, which are based on one of the most complex and derived characters in animal architecture, seem to be very attractive, these might not be the most logical ones to discuss in the context of the first metazoan animals. The presence of very basic characters, like a basal membrane or an extracellular matrix, could be argued to be more relevant in terms of evolution. If such basic characters were examined, then Placozoa probably would be inferred as the SOM in most parsimony based scenarios (See Schierwater et al. 2009). in the context of likelihood (and Bayesian) analyses, the choice of the applied models will often play a major role in methodological discussions (Yang et al. 1994; Lewis 2001a; Fan et al. 2011; Xie et al. 2011; Brown 2014; Duchêne et al. 2017; Oaks et al. 2019; Stamatakis 2019). It has recently been argued, though, that researchers can simply use the most parameter-rich model, that is, GTR+I+G (Abadi et al. 2019). Either way, discussions about integrating morphology and molecules in phylogenetic analysis often times boil down to character weighting of the two partitions, a subject that has been (Wheeler 1986; Goloboff 1993; Chippindale and Wiens 1994) and remains controversial (Goloboff et al. 2008; Simmons and Goloboff 2013; Mirande 2016; Schierwater et al. 2016; Mirande 2019). The only approach to “even out” a swamping effect that molecular characters might have over morphological characters in a combined analysis is weighting (Giribet 2010). Character weighting in simultaneous analysis of morphology and molecules is complicated by the methods that are used to collect the character information and also by the differences in the very nature of morphological versus sequence data.
It appears to us that systematists largely follow opposing trends: In molecular systematics, the data will often influence the way that characters are analyzed (through choosing appropriate models in likelihood and Bayesian approaches); whereas in morphological systematics, the method of analysis (parsimony) often seems to influence the scoring of characters. This seems to be related to fundamental differences in the two data types. Morphological characters are based on a researcher’s qualitative interpretation of complex phenotypic traits and on a subjective understanding of the morphology of the organisms being studied (Farris 1983). Molecular data, on the other hand—and phylogenomic data in particular—are to a higher extent a mixture of data with different characteristics: uninformative and informative sites, variant and invariant sites, and sites that are consistent and inconsistent with each other. Morphological data can also have these mixed features, but are typically highly curated. The Tables below exemplify this trend:
In 2017, there were several genome level analyses of the SOM as detailed in the table below. Two major contenders for SOM are CT (Ctenophores) and PO (Porifera).
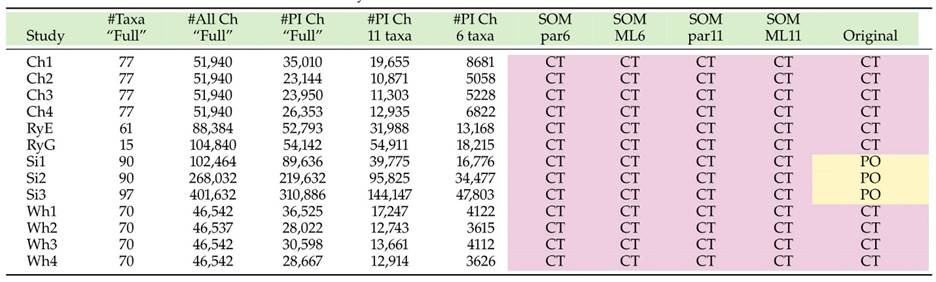
Morphological hypotheses of relationships from different studies (the matrices used are included as an appendix to this database). Two candidates for SOM arise from these analyses – PL (placozoans) and PO (poriferans). Taken from [24].
VI. TAXONOMY
Recent advances have been made in organizing the taxonomy of placozoans based on whole genome data [14] while a first system for the recognition of phylogenetic or taxonomic relatedness and diversity was developed using 16S mtDNA haplotypes [5-7]. Figure 4 summarizes our current understanding of placozoan taxonomy.
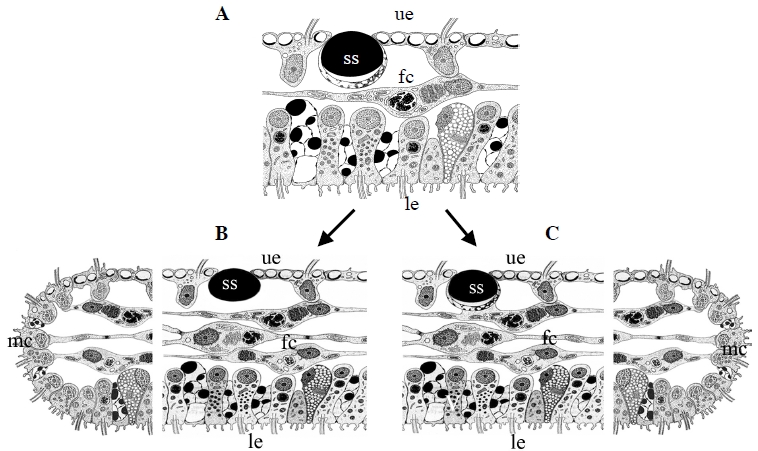
VII. DISTRIBUTION
Much work and collection has been done to decipher the global patterns of diversity. Worldwide sampling and genotyping (using the 16S mitochondrial marker) were used to determine genetic lineages and their worldwide distribution. The last major diversity study on placozoans was accomplished in 2013 [7] when ten new sites could be added to our map of placozoan distribution (Figure 5).
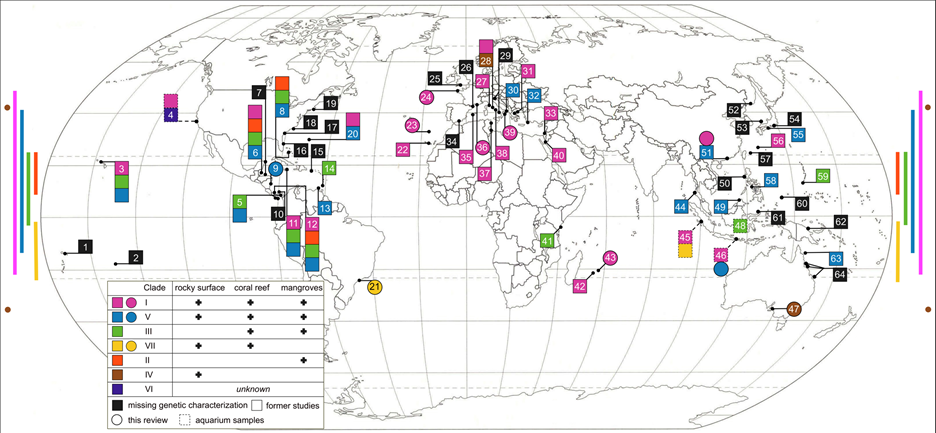
Our current knowledge of placozoan distribution also enabled further predictions on habitat suitability and distribution (Figure 6). These predictions suggest that placozoans occur in our oceans from 55°N and 44°S degrees latitude (from Northern Scotland to New Zealand) [25]. Nevertheless, species diversity and occurrence clearly appear to be greater in tropical to temperate regions. Nevertheless, species diversity and abundance clearly appears to be greater in tropical and subtropical regions.
VIII. BACTERIAL ENDOSYMBIONTS
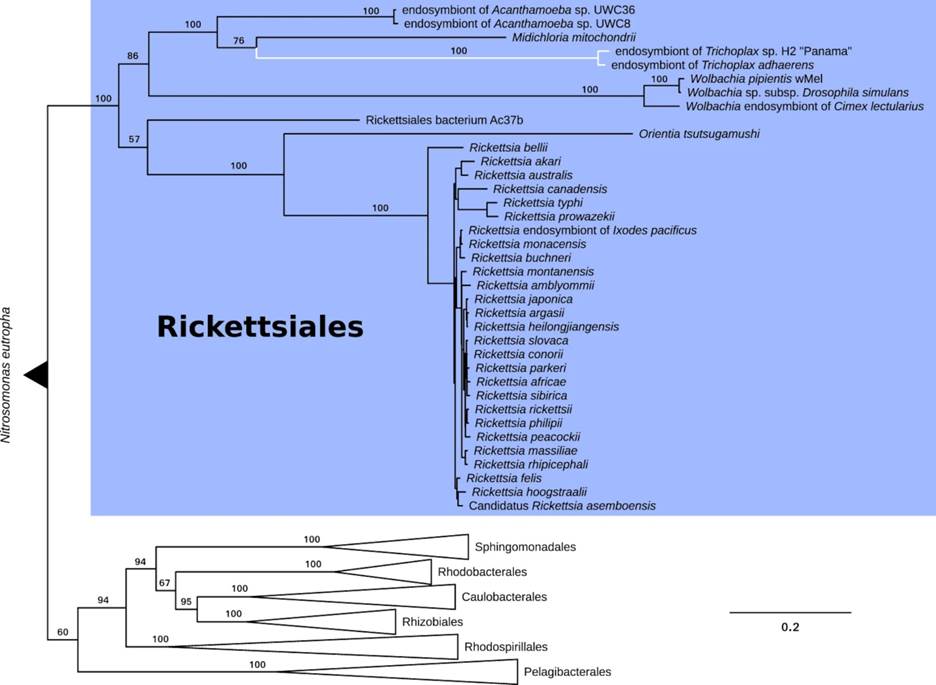
The presence of intracellular bacteria in placozoans has been reported many times, but questions remain about the true nature of this relationship [4, 16, 18, 19, 26-30]. The identified bacteria belong to the order Rickettsiales of the α-Proteobacteria (Figure 7) and related bacteria have been identified in a wide variety of marine invertebrates including stony corals. Relationships of the rickettsiales bacteria may range from pathogenic to parasitism or mutualism, depending on the bacterial species and the animal host (e.g. [30]). The relationship might even be dynamic, depending on the environmental context. For example, in stony corals, the rickettsial endosymbionts appear to be tolerable parasites but may become pathogenic [30]. In placozoans, the genetic evidence rather suggests that the symbiotic bacteria exert some form of energy parasitism (ATP translocation) but in turn may provide the host with essential amino acids and important cofactors, which in sum points to a mutualistic relationship [28].
IX. FURTHER MILESTONES IN PLACOZOAN RESEARCH
Placozoans are tiny and the simplest organized metazoans, but it appears that they have a sophisticated system of innate immunity, combined of ancestral as well as placozoan-specific components, that is able to protect them from pathogens and to manage their associated microbiota [31].
Recent findings in Placozoa, in animals without nerve cells, suggests that key components of neuronal signaling evolved prior to the invention of nerve cells in the context of paracrine cell signaling, which these animals use to coordinate movement and behavior [32].
X. SAMPLING AND ARCHIVING
Animal sampling
To sample placozoans we use two different methods (Figure 8). In the first “rock sampling” method, stones and other hard substrates, such as coral parts and mussel shells are collected at a depth of up to 3 m and placed in plastic bottles with seawater from the sampling site. As a second method, standard microscopic glass slides (76 x 26 mm) are placed in plastic microscope slide containers (“slide samples”), which are cut open at the top and the bottom to enable the flow-through of seawater [6, 7, 33]. A good, yet underestimated source for collecting placozoans are aquaria. Despite the missing exact geographic assignment of these samples it is obvious, however, that they are a reasonable sources for placozoan specimens that are at least helpful for screening genetic diversity in Placozoa.

DNA/RNA isolation
To quickly fix animals in the field for DNA preparation, FTA Elute cards micro (Whatman) are used [6, 34]. Animals are dropped onto the cards with as little seawater as possible. After drying the DNA can be stored on the card for several months to years. Alternatively, animals can be put directly in 80-98% ethanol and stored for standard DNA preparation at 4°C for several months. To preserve RNA, animals can be fixed in RNAlater (Quiagen) and processed when returning from the field to the laboratory.
Experimental studies
Placozoa are easily amenable to experimental studies, and almost the complete spectrum of biological studies has been applied to this group (see [21-23] for refs.).
New samples, new species
For the genetic identification of placozoan samples please send samples to the Schierwater lab for free genetic analysis. If new species are identified a joint effort taxonomic circle approach for valid species descriptions is encouraged (please contact Bernd Schierwater).
XI. REFERENCES
- Schulze FE: Trichoplax adhaerens, nov. gen., nov. spec. Zoologischer Anzeiger 1883, 6:92-97.
- Monticelli F: Treptoplax reptans ng, n. sp. Atti dell’Academia dei Lincei, Rendiconti (5) II 1893:39-40.
- Grell KG: Trichoplax adhaerens F.E. Schulze und die Entstehung der Metazoan. Naturwissenschaftliche Rundschau 1971, 24(4):160-161.
- Grell KG: Eibildung und Furchung von Trichoplax adhaerens F.E. Schulze (Placozoa). Z Morph Tiere 1972, 73:297-314.
- Voigt O, Collins AG, Pearse VB, Pearse JS, Ender A, Hadrys H, Schierwater B: Placozoa -- no longer a phylum of one. Curr Biol 2004, 14(22):R944-945.
- Eitel M, Schierwater B: The phylogeography of the Placozoa suggests a taxon-rich phylum in tropical and subtropical waters. Mol Ecol 2010, 19(11):2315-2327.
- Eitel M, Osigus HJ, DeSalle R, Schierwater B: Global diversity of the Placozoa. PLoS One 2013, 8(4):e57131.
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML et al: The Trichoplax genome and the nature of placozoans. Nature 2008, 454(7207):955-960.
- Kamm K, Osigus HJ, Stadler PF, DeSalle R, Schierwater B: Trichoplax genomes reveal profound admixture and suggest stable wild populations without bisexual reproduction. Sci Rep 2018, 8(1):11168.
- Eitel M, Francis WR, Varoqueaux F, Daraspe J, Osigus HJ, Krebs S, Vargas S, Blum H, Williams GA, Schierwater B, Worheide G: Comparative genomics and the nature of placozoan species. PLoS Biol 2018, 16(7):e2005359.
- Laumer CE, Gruber-Vodicka H, Hadfield MG, Pearse VB, Riesgo A, Marioni JC, Giribet G: Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias. Elife 2018, 7.
- Osigus H-J, Rolfes S, Herzog R, Kamm K, Schierwater B: Polyplacotoma mediterranea is a new ramified placozoan species. Current Biology 2019, 29(5):R148-R149.
- Miyazawa H, Osigus HJ, Rolfes S, Kamm K, Schierwater B, Nakano H: Mitochondrial Genome Evolution of Placozoans: Gene Rearrangements and Repeat Expansions. Genome Biol Evol 2021, 13(1).
- Tessler M, Neumann JS, Kamm K, Osigus H-J, Eshel G, Narechania A, Burns JA, DeSalle R, Schierwater B: Phylogenomics and the first higher taxonomy of Placozoa, an ancient and enigmatic animal phylum. Frontiers in Ecology and Evolution 2022, 10:1016357.
- Eitel M, Osigus H-J, Brenzinger B, Wörheide G: Beauty in the beast – Placozoan biodiversity explored through molluscan predator genomics. Ecology and Evolution 2024, 14 (4):e11220.
- Grell KG, Benwitz G: Die Ultrastruktur von Trichoplax adhaerens F.E. Schulze. Cytobiologie1971, 4:216-240.
- Ivanov AY: Trichoplax adhaerens, a Phagocytella-like animal. Zoologiceskij Zurnal 1973, 52:1117-1131.
- Grell KG, Benwitz G: Spezifische Verbindungsstrukturen der Faserzellen von Trichoplax adhaerens F.E. Schulze.Zeitschrift für Naturforschung C 1974, 29(11-12):790.
- Grell KG, Benwitz G: Ergänzende Untersuchungen zur Ultrastruktur von Trichoplax adhaerens F.E. Schulze (Placozoa). Zoomorphology 1981, 98:47-67.
- Guidi L, Eitel M, Cesarini E, Schierwater B, Balsamo M: Ultrastructural analyses support different morphological lineages in the phylum placozoa Grell, 1971. Journal of Morphology 2011, 272(3):371-378.
- Schierwater B: My favorite animal, Trichoplax adhaerens. BioEssays 2005, 27(12):1294-1302.
- Schierwater B, Osigus HJ, Bergmann T, Blackstone NW, Hadrys H, Hauslage J, Humbert PO, Kamm K, Kvansakul M, Wysocki K, DeSalle R: The enigmatic Placozoa part 2: Exploring evolutionary controversies and promising questions on earth and in space. BioEssays 2021, 43(10):2100083.
- Schierwater B, Osigus HJ, Bergmann T, Blackstone NW, Hadrys H, Hauslage J, Humbert PO, Kamm K, Kvansakul M, Wysocki K, DeSalle R: The enigmatic Placozoa part 1: Exploring evolutionary controversies and poor ecological knowledge. BioEssays 2021, 43(10):2100080.
- Neumann JS, Desalle R, Narechania A, Schierwater B, Tessler M: Morphological Characters Can bly Influence Early Animal Relationships Inferred from Phylogenomic Data Sets. Systematic Biology 2021, 70(2):360-375.
- Paknia O, Schierwater B: Global Habitat Suitability and Ecological Niche Separation in the Phylum Placozoa. PLOS ONE 2015, 10(11):e0140162.
- Grell KG, Benwitz G: Elektronenmikroskopische Beobachtungen über das Wachstum der Eizelle und die Bildung der “Befruchtungsmembran” von Trichoplax adhaerens F.E. Schulze (Placozoa). Z Morph Tiere 1974, 79:295-310.
- Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW: Bacterial DNA sifted from the Trichoplax adhaerens (Animalia: Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol Evol 2013, 5(4):621-645.
- Kamm K, Osigus HJ, Stadler PF, DeSalle R, Schierwater B: Genome analyses of a placozoan rickettsial endosymbiont show a combination of mutualistic and parasitic traits. Sci Rep 2019, 9(1):17561.
- Gruber-Vodicka HR, Leisch N, Kleiner M, Hinzke T, Liebeke M, McFall-Ngai M, Hadfield MG, Dubilier N: Two intracellular and cell type-specific bacterial symbionts in the placozoan Trichoplax H2. Nat Microbiol 2019, 4(9):1465-1474.
- Klinges JG, Rosales SM, McMinds R, Shaver EC, Shantz AA, Peters EC, Eitel M, Worheide G, Sharp KH, Burkepile DE et al: Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J 2019, 13(12):2938-2953.
- Kamm K, Schierwater B, DeSalle R: Innate immunity in the simplest animals - placozoans. BMC Genomics 2019, 20(1):5.
- Najle SR, Grau-Bové X, Elek A, Navarrete C, Cianferoni D, Chiva C, Cañas-Armenteros D, Mallabiabarrena A, Kamm K, Sabidó E et al: Stepwise emergence of the neuronal gene expression program in early animal evolution. Cell 2023, 186(21):4676-4693.e4629.
- Pearse VB, Voigt O: Field biology of placozoans (Trichoplax): distribution, diversity, biotic interactions. Integr Comp Biol 2007, 47(5):677-692.
- Signorovitch AY, Dellaporta SL, Buss LW: Caribbean placozoan phylogeography. Biol Bull 2006, 211(2):149-156.
XII. QUICK GUIDE: PLACOZOA
Bernd Schierwater* & Robert DeSalle**
*TiHo Hannover, ITZ Ecology & Evolution, Buenteweg 17d, 30559 Hannover.
**Comparative Genomics Institute at the American Museum of Natural History, 79th Street at Central Park West, New York, NY 10024.
What is Placozoa?
To us, Placozoa are the most unique animals one can think of (Figure 1). The bauplan of these always hungry metazoans is by far the simplest of all animals (only some secondarily reduced parasites are similarly simple in morphology). They are organized in a sandwich-like body plan with a lower epithelium (facing the substrate) and an upper epithelium (facing the open water). Embedded within these two layers is a loose layer of so-called fiber cells, and they lack any kind of symmetry or body axis. They have no discrete organs, nerve or muscle cells, nor even an extracellular matrix or a basal membrane. Just six somatic cell types perform all functions of holding the animal together (even in heavy wave break zones). Yet they can feed, digest, smell, see, move, grow and reproduce. They crawl as flat, tiny plates over different hard substrates in most ocean waters, hence their etiology.
How did it get its name?
There are two taxonomic etiologies to consider with this animal. The fist concerns the species name Trichoplax adhaerens and the second, the phylum name Placozoa. Trichoplax was discovered in 1883 by the German zoologist Franz Eilhard Schulze, in a seawater aquarium at the Zoological Institute in Graz, Austria. The generic name is derived from the classical Greek θρίξ (thrix), "hair", and πλάξ (plax), "plate". The specific epithet “adhaerens” comes from Latin "adherent", reflecting its propensity to stick to the glass slides and pipettes (and indeed the side of glass surface of aquaria) used in its examination. It is therefore affectionately known as the “sticky hairy plate” (gr. Τριξη; trich = hair, gr. Πλαχ; plax = plate, lat. Adhaerere = to stick). In 1971, Grell following Otto Bütschli’s “placula hypothesis” named the phylum “Placozoa” (gr. placos, gr. zóon). Bütschli’s hypothesis was erected to propose the morphology of the Urmetazoan or the common ancestor of all metazoans.
How do they reproduce?
Placozoans may use three different modes of reproduction: vegetative budding, vegetative fission and bisexual reproduction. Vegetative fission of a parent into two (sometimes three or more) daughter individuals is the dominant mode of reproduction in the lab, where it produces high propagation rates of clonal lineages. Sexual reproduction has been shown to occur in nature and has been induced in the lab, where the embryos develop only to the 128 cell stage and then die.
Where do we find them?
Originally it was believed that placozoans only occur in warm subtropical and tropical ocean waters. Now we predict that they even occur in moderately cold waters and the distribution range now goes as far as 55°N (e.g. coasts of Ireland) and 44°S (e.g. Tasmania). While we often don’t even have to take our 501s off to walk into the water and collect them, we also found them in 20 m depth. Most likely they will not go much deeper because their main diet, photosynthetic algae, would change, but we do not know yet.
Why is there so much interest in them?
Trichoplax and other placozoans represent novel model organisms with tremendous potential for many areas of biological and biomedical research. Relevant characteristics of these most simple metazoans include that they (i) show the most simple (but not secondarily reduced) of all metazoan bauplans, (ii) possess the smallest nuclear genome, (iii) possess the largest mitochondrial animal genome, (iv) harbor representatives of all major gene families known from humans, and (v) are exceptionally suited for both field work and experimental studies in the laboratory. They are also one of only five metazoan higher taxa that are involved in one of the most hotly debated phylogenetic questions since genomics came on the scene and that is which of Cnidaria, Ctenophora, Porifera,Placozoa or Bilatera harbor the “mother of all metazoans”. There are 105 ways that five taxa can be arranged in strictly bifurcating trees and nearly a quarter of these have been offered as hypotheses for the relationships of these five taxa.
Are placozoans the best living surrogate for the “Urmetazoon”?
Possibly yes. But if so, it would take a while for the community to accept this traditional view. If one only considers comparative organismal evidence and genuine biological knowledge, a basal position of Placozoa results as naturally as a baby’s smile (in sharp contrast to e.g. sponges, which many authors consider direct descendants of choanoflagellates because of a single character (and meanwhile outworn) argument). If one also adds functional morphology and evolutionary constraints to the soup, a placozoan first position seems cum grano salis indisputable. BUT! Recent DNA sequence information contradicts this traditional placula view of metazoan evolution. We live in a time of quantity over quality and of proximal over distal and so the current genome based view of metazoan phylogeny leaves the Placozoa somewhere in the middle of recently constructed trees.
Goodness me, will there be any solution to finding the right tree?
The answer has been given in the brilliant paper by Ian Tattersalll (2013).
Areplacozoansin the fieldoverlooked by researchers?
Yes, all the time.
What can we learn from them?
A lot can be learned from these tiny masters of morphological simplicity. As a rule of thumb, it is often helpful to first learn the basics before you analyze more complex entities. For example, if you want to count to 99% it is helpful to learn the numbers 0 to 9 first. Or let’s take cancer research as an example. Placozoans have all major gene families that researchers examine in the context of cancer. So why not first figure out what these genes are doing and how they interact in the simplest metazoan on the planet with just a small number of possible gene interactions instead of going immediately to the unmanageable numbers of possible combinatorial effects in mammals? There are other obvious examples of placozoan research utility. If we want to learn about the evolution and structural organization of genomes, why not start with the smallest and potentially most ancestral animal genome? Or if we want to learn about the development and evolution of nervous systems and other organs, why not first understand the basic genetics underlying neural function or epithelium formation in the least complex system?
IsTrichoplax adhaerensthe best placozoan species for experimental research?
No. The the close relative, Trichoplax sp. H2 is much more robust, much more abundant and much easier to culture and manipulate.
Any more complications?
Yes, despite placozoans being the simplest animals with the smallest of all early branching animal genomes, these animals might still be too complex and morphologically also too different for too simple conclusions. This is what makes comparative zoology so much fun.
Age of the oldest PhD student working onplacozoansat the time of thesis defense?
97 years, Heinz Wenderoth.
XIII. COOL (VERY) PHOTOGRAPHS AND DRAWINGS OF PLACOZOA
https://animaldiversity.org/accounts/Placozoa/
https://www.ecolevol.de/placozoa/
https://www.nature.com/articles/nature07191/figures/1
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0136098
https://mitocondriacientifica.blogspot.com/2017/04/placozoos-plaquitas-vivas-con-patas.html
https://thenode.biologists.com/a-day-in-the-life-of-a-trichoplax-lab/lablife/
Citation
Usage of data from the World Placozoa Database in scientific publications should be acknowledged by citing as follows:- Schierwater, B.; Eitel, M.; Osigus, H.-J.; DeSalle, R. (2026). World Placozoa Database. Accessed at https://www.marinespecies.org/placozoa on 2026-01-18. doi:10.14284/655
Individual pages are individually authored and dated. These can be cited separately: the proper citation is provided at the bottom of each page.
